Download Breakpoint Edges CSV
Prediction
ecDNA Amplicon Predicted: Y
Predicted Amplicon Classes: ['ecDNA', 'Linear amplification', 'Complex non-cyclic']
Predicted Genes on ecDNA
['CACNA1C', 'CACNA1C-AS1', 'CACNA1C-AS2', 'CACNA1C-AS4', 'CACNA1C-IT2', 'CACNA1C-IT3', 'CACNA2D4', 'DCP1B', 'ITFG2-AS1', 'LRTM2', 'ADAM17', 'ASAP2', 'ATP6V1C2', 'CMPK2', 'CPSF3', 'CYS1', 'GRHL1', 'HPCAL1', 'IAH1', 'ID2', 'ID2-AS1', 'ITGB1BP1', 'KIDINS220', 'KLF11', 'MBOAT2', 'NOL10', 'NRIR', 'ODC1', 'ODC1-DT', 'RN7SL832P', 'RNF144A', 'RNF144A-AS1', 'RRM2', 'RSAD2', 'SNORA80B', 'TAF1B', 'YWHAQ', 'ADAM23', 'CCNYL1', 'CPO', 'CREB1', 'DYTN', 'FAM237A', 'FASTKD2', 'FZD5', 'KLF7', 'MDH1B', 'METTL21A', 'MYOSLID', 'MYOSLID-AS1', 'PLEKHM3', 'ACBD7', 'ACBD7-DCLRE1CP1', 'ACVR1B', 'AGAP2', 'AGAP2-AS1', 'ANAPC5', 'ANO6', 'APBB1IP', 'ARHGEF25', 'ARL6IP4', 'ARMC3', 'ARMC4', 'ARMC4P1', 'ATG101', 'AVIL', 'AXIN2', 'B3GNT4', 'B4GALNT1', 'BEST3', 'BMI1', 'BTG1', 'C12orf29', 'C12orf50', 'C12orf56', 'C12orf74', 'C12orf80', 'CAMK1D', 'CBX5', 'CCDC3', 'CCER1', 'CCNT1', 'CCT2', 'CD300LF', 'CDC123', 'CDK4', 'CDNF', 'CELF2-AS1', 'CELF2-AS2', 'CEP112', 'CEP290', 'CFAP54', 'CLIP1', 'CLIP1-AS1', 'CLLU1', 'CLLU1OS', 'COMMD3', 'COMMD3-BMI1', 'COPZ1', 'CPM', 'CPSF6', 'CRADD', 'CTDSP2', 'CYP27B1', 'DBX2', 'DCLRE1C', 'DCLRE1CP1', 'DCN', 'DIABLO', 'DIP2B', 'DTX3', 'EEA1', 'EEF1AKMT3', 'EPYC', 'FAM107B', 'FAM171A1', 'FAM186B', 'FAM238A', 'FLJ12825', 'FMNL3', 'FRS2', 'GNS', 'GPR84', 'GRASP', 'HCAR1', 'HCAR2', 'HCAR3', 'HMGA2', 'HMGA2-AS1', 'HSPA14', 'IL31', 'ITGA5', 'KANSL2', 'KCNH3', 'KDM2B', 'KERA', 'KIF5A', 'KITLG', 'KNTC1', 'KRT7', 'KRT7-AS', 'KRT86', 'KRT87P', 'LEMD3', 'LRRC10', 'LRRC43', 'LUM', 'LYZ', 'MARCHF9', 'MCM10', 'MCRS1', 'MDM2', 'MEIG1', 'METTL1', 'MKRN9P', 'MLLT10', 'MRC1', 'MRPL42', 'MSRB3', 'MYRFL', 'NAB2', 'NACA', 'NCKAP1L', 'NEBL', 'NEDD1', 'NELL2', 'NMT2', 'NR4A1', 'NRP1', 'NUDT4', 'NUDT5', 'NUP107', 'OLAH', 'OPTN', 'OR6C1', 'OR6C3', 'OR6C6', 'OR7E47P', 'OS9', 'PARD3', 'PDE1B', 'PHYH', 'PIP4K2A', 'PIP4K2C', 'PITPNM2', 'PLEKHA8P1', 'PLEKHG7', 'PPIAP30', 'PRIM1', 'PRPF40B', 'PTCHD3', 'RAB37', 'RAB3IP', 'RACGAP1P', 'RAP1B', 'RGS9', 'RNF34', 'RPP38', 'RPP38-DT', 'RPSAP52', 'RSRC2', 'SCAT2', 'SEC61A2', 'SEPHS1', 'SFTA1P', 'SKIDA1', 'SLC16A7', 'SLC26A10', 'SLC35E3', 'SLC4A8', 'SMIM41', 'SMUG1', 'SNORA113', 'SNORA2A', 'SNORA2B', 'SNORA2C', 'SNORA70G', 'SPAG6', 'SPATS2', 'ST8SIA6', 'STAM', 'STAM-AS1', 'STAT6', 'SUV39H2', 'TBC1D30', 'TMBIM6', 'TMEM117', 'TMTC3', 'TSFM', 'TSPAN31', 'UBE2N', 'UCMA', 'USP6NL', 'VPS33A', 'VPS37B', 'WIF1', 'YEATS4', 'ZCCHC8', 'ZNF385A']
Predicted Features
View Feature 1 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
10 |
0.6794647768 |
View Feature 2 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
11 |
0.6930890471 |
View Feature 3 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
12 |
0.6902710714 |
View Feature 4 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
13 |
0.646136709 |
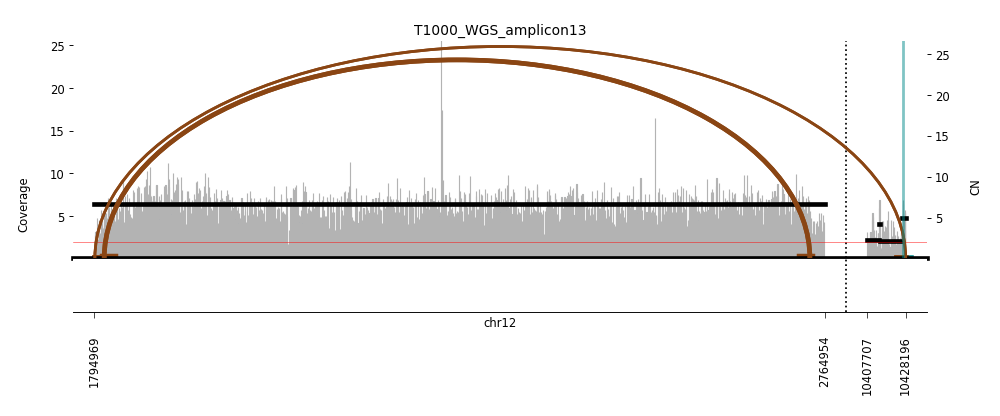
View Feature 5 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
14 |
0.0 |
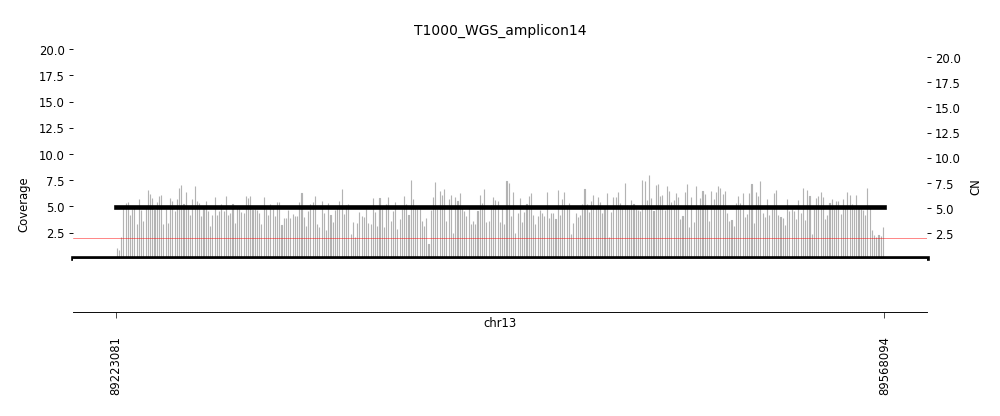
View Feature 6 - T1000_LIPOSARCOMA Complex non-cyclic
| Class |
Amplicon |
Complexity Score |
| Complex non-cyclic |
15 |
0.7625110024 |
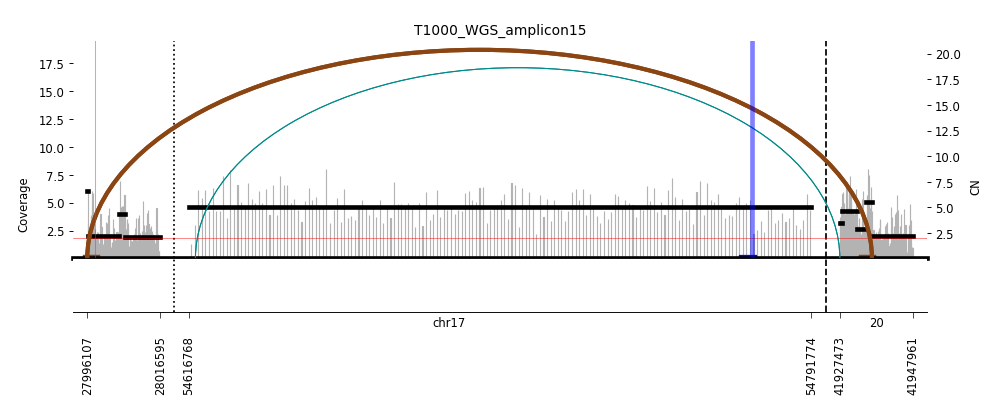
View Feature 7 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
1 |
0.6470866844 |
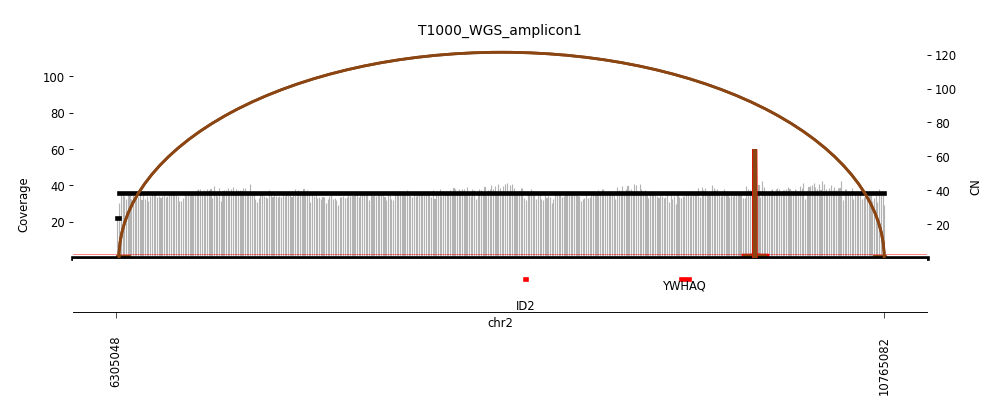
View Feature 8 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
2 |
0.0 |
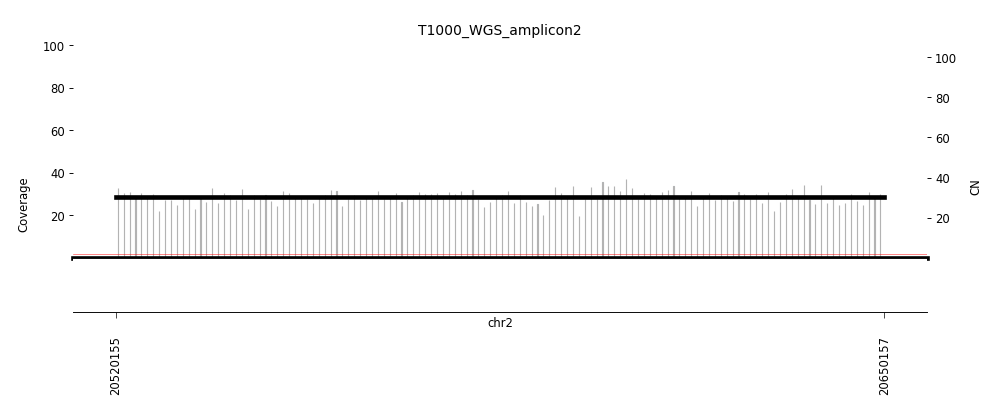
View Feature 9 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
3 |
0.8043779375 |
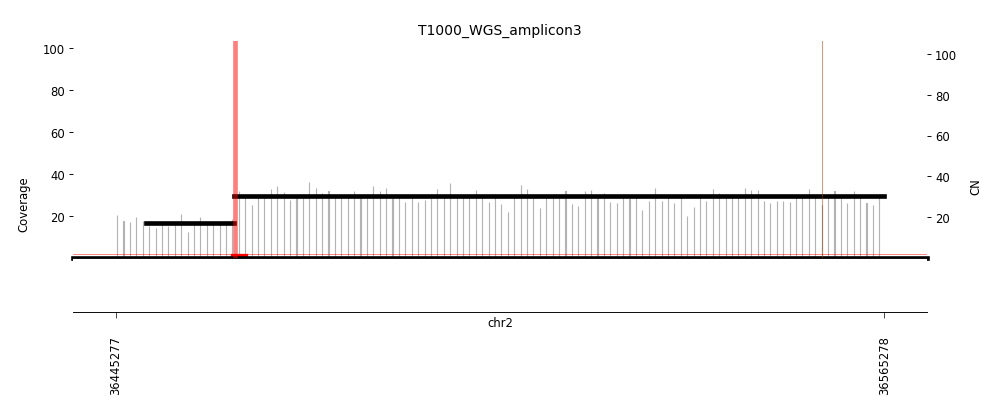
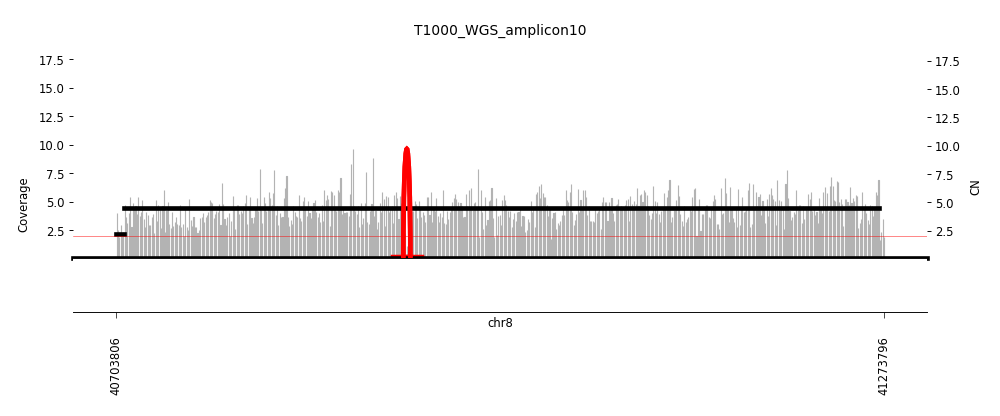
View Feature 10 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
4 |
0.0 |
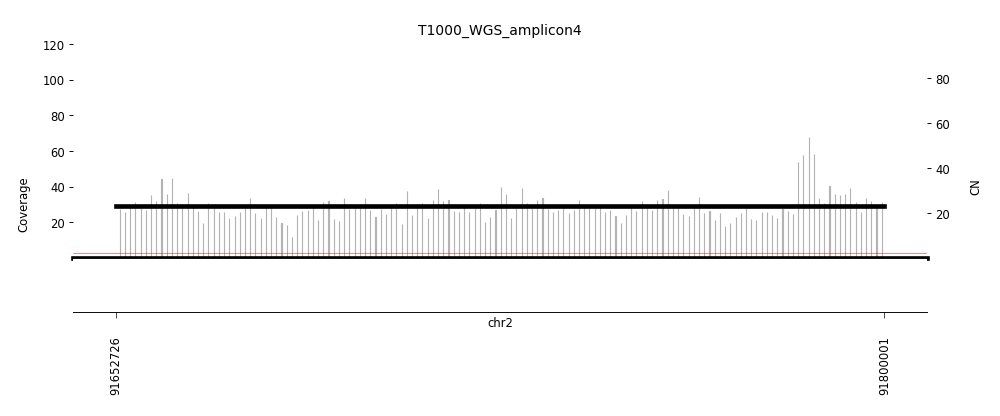
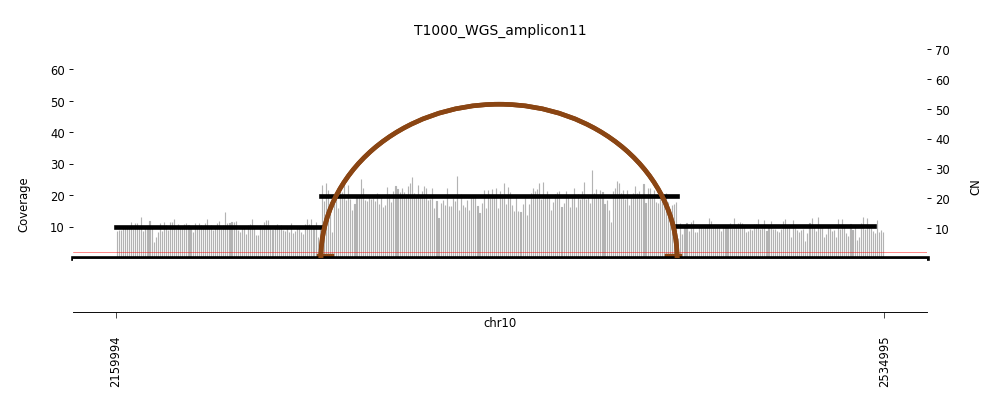
View Feature 11 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
5 |
0.607012619 |
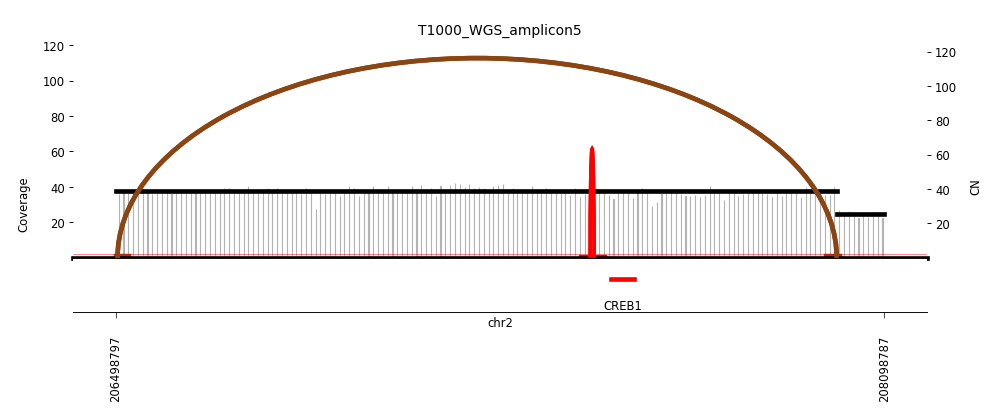
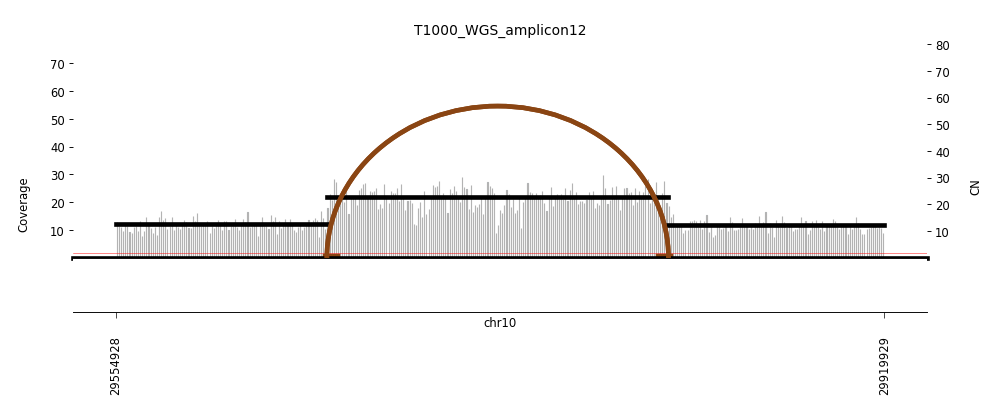
View Feature 12 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
6 |
0.6297306158 |
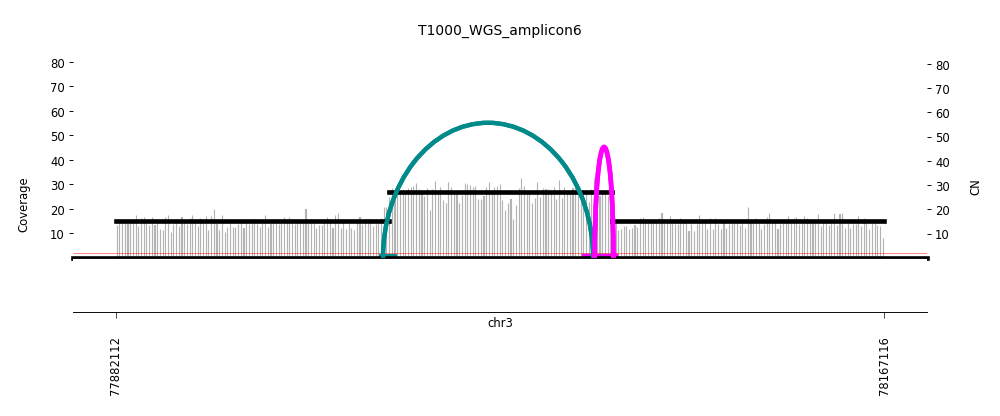
View Feature 13 - T1000_LIPOSARCOMA ecDNA
| Class |
Amplicon |
Complexity Score |
| ecDNA |
8 |
3.052346652 |

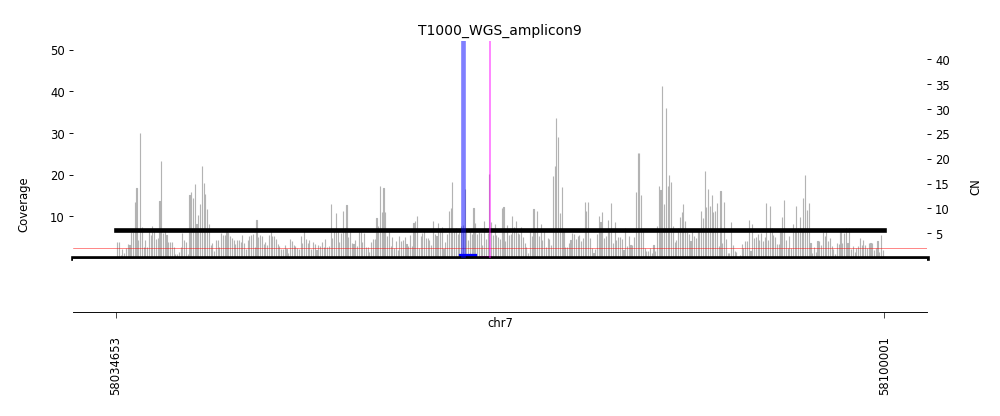
View Feature 14 - T1000_LIPOSARCOMA Linear amplification
| Class |
Amplicon |
Complexity Score |
| Linear amplification |
9 |
0.4528621554 |